- Home
- Research Areas
- Research Spotlight: Predictive Biology

Using HPC to develop predictive analytics that will expedite the characterization of biological threats, predict the paths of diseases through the population, and accelerate the development of technologies and treatments
Originating as a collaboration called Biological Applications of Advanced Strategic Computing (BAASIC), LLNL and its partners—other Department of Energy (DOE) laboratories, government agencies, academic institutions, and industry members—apply high-performance computing (HPC) to biological research. From virtual screenings that identify the most promising drug candidates to visualization of the brain’s neural connections, these computational predictive biology research efforts have helped make the world a healthier and safer place by integrating supercomputing, data analysis, and predictive simulations at scale.
Research Applications
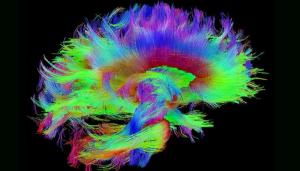
Traumatic brain injuries (TBIs) affect millions of people each year, whether from car accidents, sports injuries, or on the battlefield. The Defense and Veterans Brain Injury Center reported nearly 414,000 TBIs among U.S. service members worldwide between 2000 and late 2019—yet treatment and, in many cases, even diagnosis remain elusive. Making sense of the brain with its 100 billion neurons and tens of thousands of connections in the service of veterans is perhaps one of the best possible applications of data science and the use of LLNL’s expertise in data science expertise and high-performance computing (HPC).
LLNL’s involvement in TBI research sprang from discussions held in 2019 involving several national laboratories, the Department of Veterans Affairs, and the University of California, San Francisco, with a goal of bringing precision diagnostics and prognostic modeling to TBI by sifting through a large and complicated dataset to form a picture of TBI that eventually can be applied in a clinical setting. Read more about the Lab’s investigation of traumatic brain injury, including the TRACK-TBI project and the MaPPeRTrac brain tractography workflow.

Several national laboratories—LLNL, Los Alamos, Argonne, and Oak Ridge—have teamed up with the National Cancer Institute (NCI) for a three-part pilot program designed to integrate supercomputing into cancer patient treatments. The program stretches the scientific frontiers of supercomputing and lays the foundation for the next major advances in cancer therapy.
Pilot 1 (Analyzing Drug Effectiveness through Advanced Computing) develops a library of validated prediction tools for projecting drug screen results and improving the efficacy of drug selection for patients.
Pilot 2 (Targeted Cancer Therapy using Innovative Molecular Dynamics Simulations at Unprecedented Scales) helps improve health and treat disease by enabling a fundamental understanding of the RAS protein’s activation and predicting the success of new oncology therapies. Read more about the Lab's multiscale modeling for cancer and the Massively parallel Multiscale Machine-Learned Modeling Infrastructure (MuMMI) framework, which is part of the ADMIRRAL (AI-Driven Multiscale Investigation of RAS-RAF Activation Lifecycle) project. Through these initiatives, researchers hope to gain an understanding of how the RAS and RAF proteins mutate and cause tumors to form.
Pilot 3 (Improving Cancer Care with Computer Modeling) improves cancer care by applying advanced computation to population-based cancer data in order to understand the impact of new diagnostics, treatments, and patient factors in real-world patients.

From the outset of the COVID-19 pandemic, LLNL has been fully committed to helping protect the U.S. from the virus and to speed the recovery of those affected. As one effort, a public data portal provides a wealth of data LLNL scientists have gathered from their ongoing COVID-19 molecular design projects, particularly the computer-based “virtual” screening of small molecules and designed antibodies for interactions with the SARS-CoV-2 virus for drug design purposes. The data is queryable by criteria such as chemical structure and binding probability scores, so outside researchers can easily locate relevant data for their own work. The portal is regularly updated and will provide the results of experiments performed at the Laboratory on the effectiveness of small molecules and antibodies against SARS-CoV-2.
Additionally, an interactive visualization tool includes drug compounds and therapeutic agents mentioned in the COVID-19 Open Research Dataset and extracted using natural language processing. This interactive data visualization dashboard allows researchers to explore which drug compounds have been used to treat and/or study COVID-19. Researchers studying treatments for COVID-19 can quickly visualize the drug compounds that have been studied, the combination of drugs that have been explored, and the classes of drugs that are being mentioned in scientific literature.

Pharmaceutical companies are looking for faster, more accurate ways to identify promising drug compounds and evaluate the efficacy and safety of new drugs. Such a capability could reduce costs and risks, thus allowing companies to bring antibiotics and other drugs more quickly to market. The solution may lie in supercomputer-based modeling and simulation. LLNL researchers are collaborating with several companies in this pursuit.
For example, in a partnership with the American Heart Association (AHA), LLNL researchers have been working to overcome the burden of drug discovery, cost and access. The two organizations created a simulated environment that rapidly and precisely predicts how drugs bind to their target proteins. These predictions were tested in experimental systems to generate a robust drug pipeline of new and targeted therapies.
LLNL is also part of the ATOM Consortium (Accelerating Therapeutics for Opportunities in Medicine), a public-private collaboration that aims to cut preclinical cancer drug discovery from six years to just one. ATOM is developing, testing, and validating a multidisciplinary approach to drug discovery in which modern science, supercomputing simulations, data science, and artificial intelligence are highly integrated into a single drug-discovery platform that can ultimately be shared with the drug development community at large. Read more about the Lab's data-driven pharmaceutical models.





